Recently, The X-Gen AI New Drug Discovery and Design Research Center has published a paper titled" Dual Computational and Experimental Strategy to Enhance TSLP Antibody Affinity for Improved Asthma Treatment" in PLOS Computational Biology, a high-impact classic journal in the field of bioinformatics and computational Biology. (doi: 10.1371 / journal. Pcbi. 1011984). Jointly trained doctoral student Lv Yitong from X-Gen AI New Drug Discovery and Design Research Center is the first author. Researcher Xu Lida is the main corresponding author. Professor Yu Changyuan of Beijing University of Chemical Technology is the co-corresponding author, and General manager Sun Zhiwei of Sungen Biomedical provided extremely important guidance for the construction of research processes.
The study establishes a route that helps antibody affinity maturation through computation, effectively predicts key amino acids, and uses these predictions to guide directed mutations of antibodies. The team successfully obtained a high-affinity antibody targeting TSLP, effectively shortening the antibody affinity maturation time. The results were agreed by the reviewers. "This research has important implications for promoting the development of targeted therapeutic antibodies". Comments of the paper are also published on the journal's website.

The cytokine thymic stromal lymphopoietin (TSLP), derived from epithelial cells, is involved in the initiation and persistence of asthma inflammatory pathways. It has been found that TSLP forms a trimeric signaling complex with the thymic stromal lymphopoietin receptor (TSLPR) and Interleukin-7 receptor alpha chain (IL-7Rα), activating intracellular signaling via the STAT5 pathway, which leads to the release of inflammatory cytokines. Targeted antibody drugs against TSLP and its signaling pathway are considered effective strategies for asthma treatment. They screened fully synthetic human phage antibody libraries and successfully identified T6, a specific antibody that targets TSLP. Subsequently, they employed experimental alanine scanning to identify critical amino acids, ensuring the accuracy of the T6-TSLP complex structure.
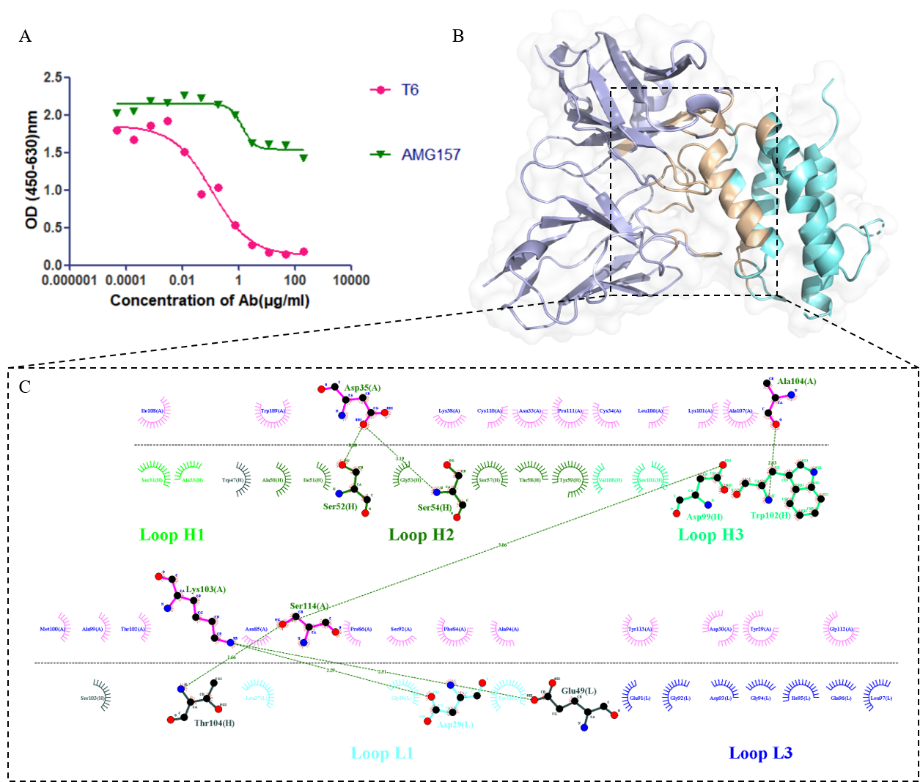
Fig.1: Structure and interaction of the TSLP-T6 complex. (A) ELISA experiments determining the binding epitopes of AMG157 and T6 on TSLP. (B) Complex structure of TSLP and T6. (C) Ligplot+ analysis of the interactions between TSLP and T6.
Computational tools were used to predict the effect of key amino acid mutations on antibody-antigen binding affinity, and mutations with potential affinity enhancement were selected for experimental verification. Finally, after two rounds of mutation, the team succeeded in improving the affinity of a TSLP antibody, better than antibody AMG157 on the market. This set of processes can effectively shorten the time of antibody affinity maturation.
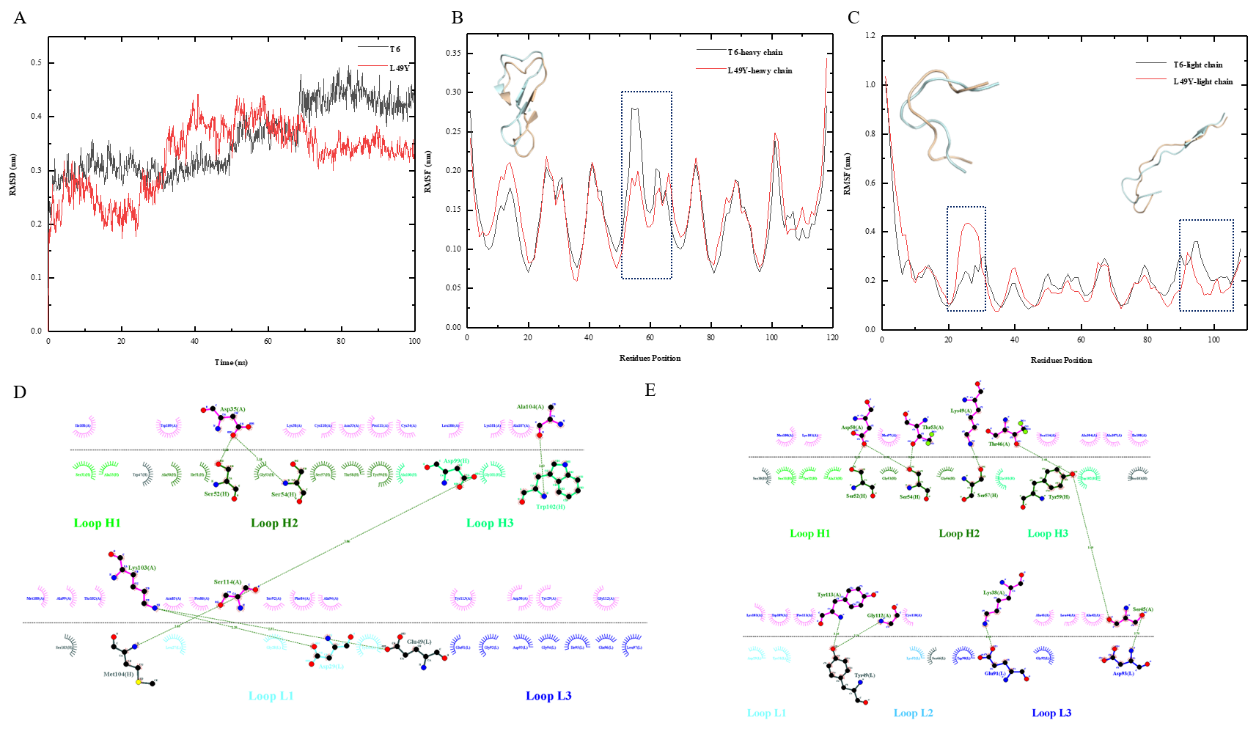
Fig.2: Dynamic simulation and force analysis graphs. (A) RMSD plot of the L49Y dynamic simulation; (B) RMSF plot of the L49Y heavy chain dynamic simulation; (C) RMSF plot of the L49Y light chain dynamic simulation; (D) Force analysis plot of the T6-TSLP interaction; (E) Force analysis plot of the L49Y-TSLP interaction.
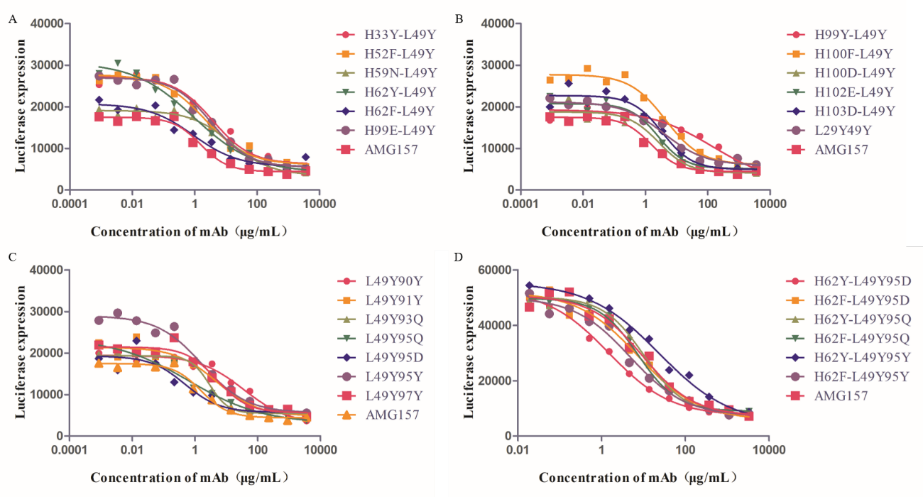
Fig.3: Antibody blocking efficacy.
Findings demonstrate the potential of computer-assisted techniques in expediting antibody affinity maturation, thereby reducing both the time and cost of experiments. The integration of computational methods with experimental approaches holds great promise for the development of targeted therapeutic antibodies for TSLP-related diseases.
Updated: Jul 24, 2024